
Dr. L | Feb, 28 2020
The recent emergence of Coronavirus Disease in 2019 (COVID-19), caused by infection by SARS-CoV-2, has put the world on high alert. People are concerned about how soon drugs and vaccines will be available after the isolation of virus strains and the completion of sequencing and comparative analysis. The scientific community is also dedicated to researching the mechanism of viral infection, with the aim to achieving precise control as soon as possible.
Coronavirus spike (S) glycoprotein and receptor ACE2 are key binding sites for severe acute respiratory syndrome coronavirus (SARS-CoV). Due to the similarity of the genetic sequences and pathogenesis between SARS-CoV-2 and SARS-CoV, the scientific community has once again turned its attention to the study of coronavirus spike and receptor ACE2 interactions. The analysis of the binding site of the S protein and the host receptor protein is of great significance, allowing us to better understand the mode of viral invasion and its mechanism of action (MOA) on the receptor. This information is not only critical for the identification of strategies to prevent further spread of SARS-CoV-2, but also serves as guidance for preventing and controlling the emergence of coronaviruses in the future.
1. Domain analysis of the Coronavirus (CoV) spike (S) protein
The key determinant of CoV host specificity is the spike (S) glycoprotein, which can be further divided into a S1 subunit and a S2 region. In general, the S protein initiates infection by mediating receptor-recognition and membrane fusion - the mechanism of which can be concluded as follows:
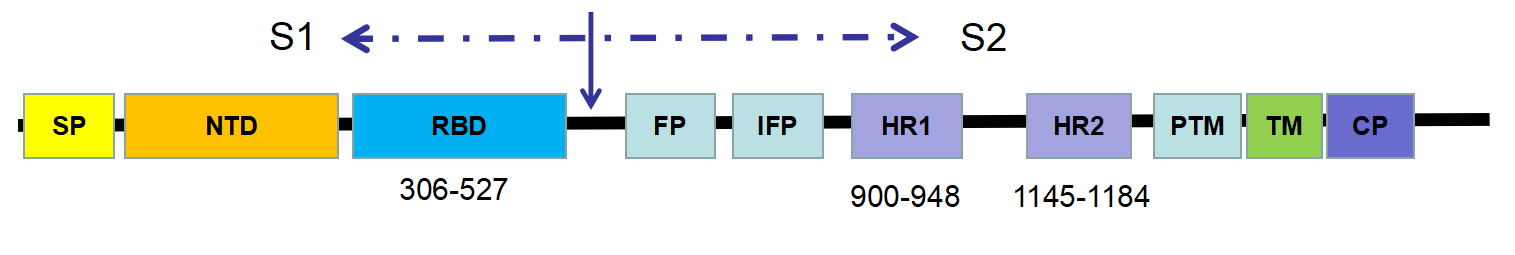
Figure 1- Schematic representation of the SARS-CoV spike (S) protein
Although SARS-CoV-2 and SARS-CoV fall within different clades, they still possess around 50 conserved amino acids in S1. It suggests that the structure of SARS-CoV-2 is closed to that of SARS-CoV. Furthermore, both SARS-CoV and SARS-CoV-2 belong to the β category of coronaviruses – for which the receptor-binding domain is commonly located in the C-terminal domain of S1 (figure 2). Hence, it can be estimated that the host receptor of SARS-CoV-2 might be similar to that of SARS-CoV.
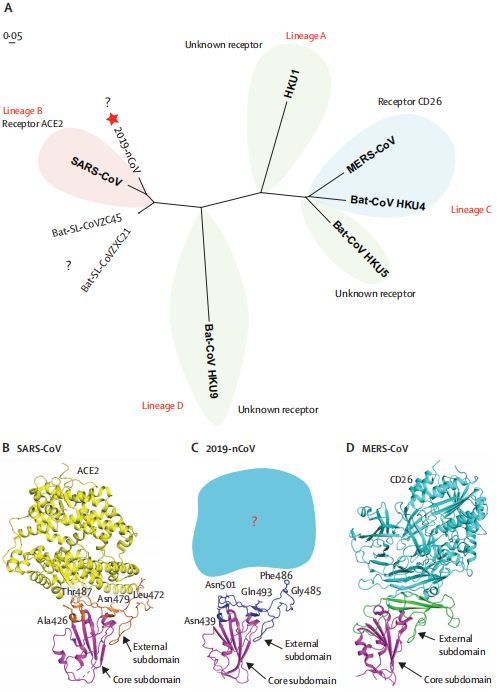
Figure 2- Phylogenetic analysis and homology modelling of the receptor-binding domain (RBD) of
the SARS-CoV-2,SARS-CoV, and MERS-CoV (Roujian Lu, 2020)
(A) Phylogenetic analysis of the receptor-binding domain from various β coronaviruses.
The star highlights SARS-CoV-2 and the question marks means that the receptor used by the viruses remains unknown. Structural comparison of the receptor-binding domain of SARS-CoV (B), SARS-CoV-2 (C), and MERS-CoV (D) binding to their own receptors. Core subdomains are magenta, and the external subdomains of SARS-CoV, SARS-CoV-2, and MERS CoV are orange, dark blue, and green, respectively. Variable residues between SARS-CoV and SARS-CoV-2 in the receptor-binding site are noted by sticks. (2019-nCoV: 2019 novel coronavirus; MERS: Middle East respiratory syndrome coronavirus.)
To enter host cells, SARS-CoV needs to first bind to the ACE2 cell-surface receptor via the viral receptor binding domain (RBD). Based on the sequence similarities between SARS-CoV-2 and SARS-CoV, the possibility for them to share the same receptor (ACE2) can be predicted. Previous studies show that ACE2, expressed in HeLa cell lines, can be used as the receptor of SARS-CoV-2. However, the binding mechanism remains unclear.
2. Structural analysis of ACE2 receptor protein
ACE2 is a type I membrane glycoprotein and contains a large N-terminal ectodomain built of two α-helical lobes. The complex structure of SARS-CoV RBD bound to ACE2 revealed that the viral RBD utilizes its external subdomain to exclusively engage the N-terminal lobe of the receptor (figure 3). Residues 424-494 are also referred to as the receptor binding motif (RBM), which cradle the most N-terminal helix in ACE2. Additionally, the two ridges of the RBM further interact with the receptor. The buried surface area upon complex formation is 927.8A˚2 in the SARS-CoV RBD and 884.7A˚2 in ACE2, respectively. The interface involves at least 18 residues in the receptor and 14 residues in RBD, forming a network of hydrophilic contacts that are suggested to predominate the RBD/ACE2 interactions. After binding to ACE2, fusion between the SARS-CoV envelope and the host cell membrane is executed by the S2 subunit.
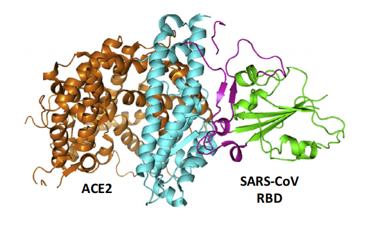
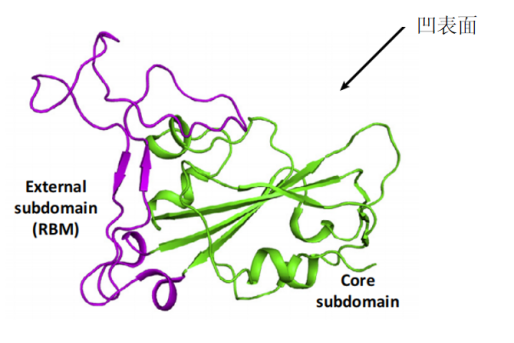
Figure 3- The complex structure between SARS-CoV RBD and its receptor, ACE2.
(Guangwen Lu, 2015)
The core (green) and external subdomains (purple) of RBD and the N- (cyan) and C-terminal lobes (orange) of ACE2 are indicated by color (noted in parentheses).
The SARS-CoV-2 strain shows around 79% sequence identity with SARS-CoV. Furthermore, it has been discovered that both the sequence of SARS-CoV-2 RBD and the interaction mechanism between RBM and ACE2 are similar to those of SARS-CoV. Thus, researchers have speculated that the ACE2 receptor is likely to be a key receptor protein for SARS-CoV-2 infection in humans.
Compared to SARS-CoV RBM, the SARS-CoV-2 RBM only differs by a one-residue insertion on a loop away from the ACE2-binding region (figure 4).
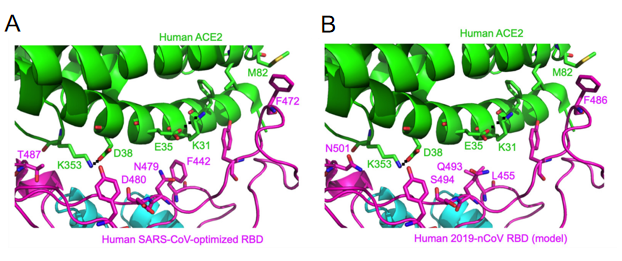
Figure 4- Structural analysis of human ACE2 recognition by SARS-CoV and SARS-CoV-2
(Yushun Wan, 2020)
Among the 14 ACE2-contacting residues in the RBD, 9 are fully conserved and 4 are partially conserved between SARS-CoV-2 and SARS-CoV.
Residue 479 in SARS-CoV RBD is located near virus-binding hotspot Lys31 (i.e., hotspot-31) on human ACE2. Hotspot-31 consists of a salt bridge between Lys31 and Glu35 buried in a hydrophobic environment. The K479N mutation removes the unfavorable interaction at the RBD/human ACE2 interface and enhances viral binding to human ACE2 - playing a critical role in the civet-to-human transmission of SARS-CoV. Importantly, Gln493 in 2019-nCoV RBD is compatible with hotspot-31, suggesting that 2019-nCoV is capable of recognizing human ACE2 and infecting human cells.
Second, residue 501 in SARS-CoV-2 RBD (corresponding to residue 487 in SARS-CoV) is an asparagine. Based on the previous structural analysis, residue 487 in SARS-CoV is located near virus-binding hotspot Lys353 (i.e., hotspot-353) on human ACE2. Hotspot-353 consists of a salt bridge between Lys353 and Asp38 also buried in a hydrophobic environment. In human SARS-CoV isolated in year 2002, residue 487 is a threonine, which strengthens the structural stability of hotspot-353. The S487T mutation adds the favorable interaction at the RBD/human ACE2 interface, enhances viral binding to human ACE2, and plays a critical role in the human-to-human transmission of SARS-CoV.
Third, residues 455, 486, and 494 are leucine, phenylalanine, and serine in SARS-CoV-2 RBD, respectively (corresponding to residues 442, 472, and 480 in SARS-CoV, respectively). Based on the previous structural analysis, these three residues in SARS-CoV RBD play significant roles, albeit not as dramatic as residues 479 and 487, in ACE2 binding.
Despite the substitution of amino acid residues, the SARS-CoV-2 spike protein still has significant binding affinity to the human ACE2 receptor. In other words, hotspot-31 and hotspot-353 may be important sites for us to study. However, mouse or rat ACE2 has histidine at 353 and it is not as efficient as lysine for the virus-receptor interaction, which provides evidence for the screening of the intermediate hosts.
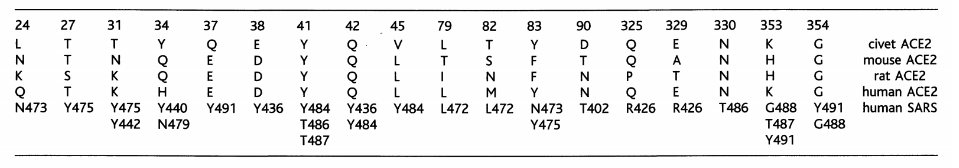
Table 1. Contacts between ACE2 in different species and SARS-CoV RBD (Fang Li, 2016)
As with both SARS-CoV and MERS-CoV, SARS-CoV-2 is a pathogen of zoonotic origin that can infect humans across barriers between species. The mechanism of virus transmission between species is an important scientific problem that needs to be solved urgently. The membrane protein of these coronaviruses recognizes the receptors and undergoes cell fusion and the spike protein needs to be cleaved by the host protein before fusion with the membrane. This process is necessary for the virus to infect the host – so blocking this process can prevent viral infection.
Conclusion
This article describes both the structure of the spike protein and the cell membrane receptor protein ACE2 as well as how they interact. Based on the previous research on SARS-CoV, the analysis of SARS-CoV-2 eventually focuses on two binding hotspots on the receptor protein: hotspot-31 and hotspot-353. To verify the relationship between these sites and clinical manifestations, in vivo studies should be conducted. In general, genetically modified mouse models are used as tools for clinical research, which might be critical for the development of vaccines and drugs.
There is still a long way to go in research efforts to combat the virus. Hopefully, this struggle between humans and microorganisms can permit us to understand our viral opponents more thoroughly.
About Cyagen
The prevention and control of coronavirus is a difficult and ongoing war. Across vaccine development, new drug screening, or gene therapy - accurate animal models are needed to verify the pathogenesis and immune mechanisms of the disease. As a leading provider of professional animal model generation services, Cyagen’s R&D department has begun development of animal models related to the research of SARS-CoV-2 since the beginning of the outbreak. Hopefully we can make a contribution to preventing and controlling this disease. In any case, we believe that we can eventually win this battle with our unity of effort.
References:
We will respond to you in 1-2 business days.