
Since the early 2000s, large-scale knockout consortia have been established with the goal of generating a complete resource of reporter-tagged null mutations in C57BL/6 mouse ES cells. Herein, we provide a brief introduction to the ‘knockout-first’ (KO-first) conditional allele strategy that forms the basis for generation of lacZ-tagged conditional alleles as part of the International Knockout Mouse Consortium (IKMC) effort organized by the International Mouse Phenotyping Consortium (IMPC). The strategies used by the IMPC have complementary properties to produce knockout alleles, but commonly “rely on the identification of a ‘critical’ exon common to all transcript variants that, when deleted, disrupts gene function.”1 Herein, we discuss the advantages provided by the KO-first allele design for elucidation of functional genomics and proteomics, as well as its contribution to generating lacZ-tagged null mutations in every protein-coding gene as part of the IMPC program.
The complete sequencing of the human and murine genomes has not only provided insights for research, but has also raised further questions regarding the understanding of gene function. The complete functional analysis of protein-coding genes remains an important goal, and technical challenge, for geneticists to-date. Some initial approaches to deciphering gene function include genome-wide mutagenesis strategies or gene trapping in mouse embryonic stem (ES) cells. Gene trapping has provided random insertional mutations in more than half of the protein-coding genes in mice, but this technique is limited by the inability to precisely engineer the gene-trap alleles, making it unsuitable for the generation of a complete set of gene knockouts. The IMPC has utilized CRISPR/Cas9 technology to generate knockout genes for several years, however, increasing concerns regarding off-target effects has cast doubts upon using CRISPR/Cas9 without thorough gene sequencing & analysis to validate the precision of the tool.
Gene targeting in embryonic stem (ES) cells remains one of the primary technologies for creating genetically modified animal models due to its accuracy and versatility. ES cell-based gene targeting has been used to establish the KO-first strategy for creating a multipurpose allele for both knockout and conditional applications by retaining the complete target gene.
The KO-first strategy has implications for functional genomics and proteomics across many model systems, including mouse, rat, and human pluripotent stem cells – serving as a functional genomics platform enabling systematic, genome-scale programs for proteomic mapping. Conditional alleles, including the KO-first allele, allow for analysis of gene function in a tissue-specific or temporal manner during development. The KO-first allele overcomes the limitations of constitutively-expressed mutations, flexibly producing reporter knockouts, conditional knockouts, and null alleles through exposure to site-specific cyclization recombination (Cre) and flippase (FLP) recombinases – such as in crosses to FLP and cre mice. Furthermore, KO-first alleles may be easily modified in ES cells using dual recombination-mediated cassette exchange. With advances in gene targeting, the KO-first allele supports “the systematic generation of doubly targeted ES cell lines for functional studies by conditional mutagenesis, which will serve to complement and extend RNA interference studies by providing complete genetic knockouts.” The spatiotemporal expression of the genes can be controlled for each study through crossing with appropriate cre mouse, even providing full-body knockouts as desired. For studies involving genes that present heterozygous lethal phenotypes, the ability to remove the FRT-flanked stop cassette is additionally useful.3
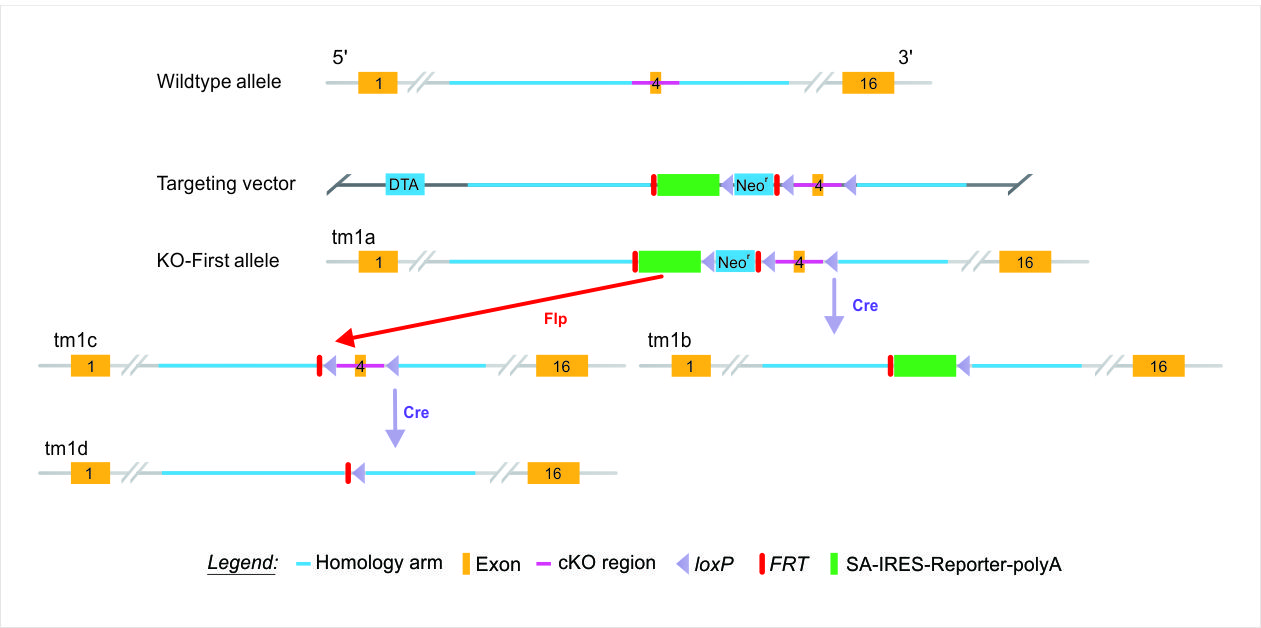
Both promoter-driven and promoterless targeting cassettes are used to generate KO-first allele constructs for EUCOMM and KOMP-CSD, as part of the IKMC effort – the initial unmodified null allele (tm1a) is achieved through “Insertion of a lacZ trapping cassette and a floxed promoter-driven neo cassette … into the intron of a gene.”1 Removal of the gene-trap cassette between FRT sites by FLP recombinase reverts the mutation to wild type, restoring gene activity, and leaves loxP sites on either side of the critical exon in the resulting conditional allele (tm1c). Exposing the conditional allele (tm1c) to Cre deletes the critical exon, inducing a frameshift mutation (tm1d allele) and triggering nonsense-mediated decay of the mutant transcript – allowing recovery of the null allele. As described, this strategy facilitates reversion of the phenotype with FLP and induction of the phenotype with Cre, which allows assessment of potential secondary linked mutations that may arise in cultured ES cells. Furthermore, mice generated with the KO-first (tm1a) allele may be crossed to an appropriate Cre driver line to remove the selection cassette and eliminate a key exon within the gene, generating the tm1b null allele – a lacZ-tagged allele for phenotyping by the IMPC. An overview of the alleles made possible with a KO-first strategy is provided below.
| Mutation | Allele | Description | Conditional Potential |
| tm1a | Knockout-First (KO-First) |
Reporter-tagged insertion allele (Null) | Yes |
| tm1b | CREed KO-First | Reporter-tagged deletion allele (for IMPC) | No |
| tm1c | FLPed KO-First | Conditional ready (restored gene activity) | Yes |
| tm1d | FLPed, CREed KO-First | Deletion allele, no reporter (Null) | No |
Table adapted from source 2.
In the last four years alone, Cyagen’s experts have provided researchers with sixteen KO-first mouse model lines for a variety of genes, including Nfe2l1, Tob1, Mir28a, Ccdc13, Tmem138, Mettl3, and more.
If you are interested in using the KO-first strategy to study your gene(s) of interest, contact us to receive a free project strategy & quote from our gene modification experts within 48 hours.
We will respond to you in 1-2 business days.