
Dr. Huang | Mar, 2 2020
At the end of 2019, a large-scale illness broke out from Wuhan, Hubei Province - a central province of China. The virus that cased this illness could enter the respiratory tract through droplets - moreover, the increasingly busy traffic conditions and travel around the holiday season made the virus spread extremely rapidly. In just over a month, the epidemic spread throughout the land of China. The viral infection can lead to an illness with symptoms including fever, difficulty breathing, impaired liver and kidney function, serious cough and pneumonia - in some severe cases, the virus can cause shock or even death.
What is the reason behind this epidemic?
The virus causing this disaster is called SARS-CoV-2, being infected by which could lead to the development of Coronavirus Disease in 2019 (COVID-19). SARS-CoV-2 falls in the β-genus of the coronavirus subfamily, alongside the viruses that causes SARS (Severe Acute Respiratory Syndrome) and MERS (Middle East Respiratory Syndrome). As implied by their names, the SARS-CoV-2 virus and SARS-CoV (from the 2002 outbreak) have closer kinship (with approximately 79% genomic sequence similarity).
In addition to the three viruses mentioned above, there are four other pathogenic coronaviruses known to infect humans. Fortunately, they have basically been able to get along with human beings peacefully - only occasionally invading humans when our immunity is reduced to cause common cold symptoms. Such common viruses have been recorded by the human immune system so they can be easily captured and eliminated by immune cells and antibodies in a timely manner, preventing an additional infection. However, the immune system had never encountered the viruses that caused SARS and MERS. Therefore, at the beginning of the virus outbreak, our bodies could only defend themselves using the varied mechanisms of human innate immune response. In this situation, viral immune escape causes some people's immune systems to either respond too slowly or overreact to attack their own cells, leading to cell death. The world was caught off guard in 2002 and 2012 due to the spreading of SARS and MERS: there were over 8,000 confirmed SARS infections in people and more than 2,000 confirmed MERS infections in people worldwide, with mortality rates of 9.6% and 35%, respectively.
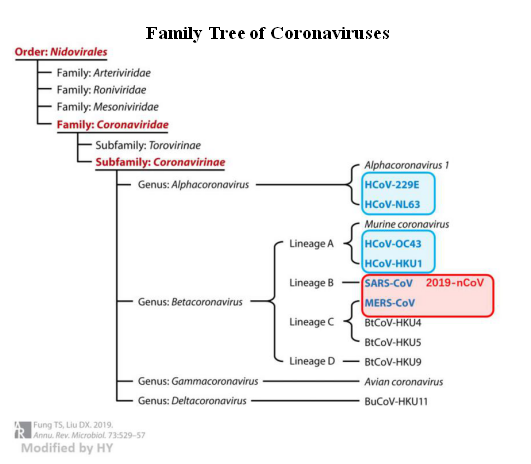
Figure 1. Human Coronavirus (Fung & Liu, 2019)
How can pathogenic coronaviruses spread from natural hosts to humans?
Five natural hosts of the seven pathogenic coronaviruses are bats, with two originating from wild rodents. Three types of coronaviruses (SARS-CoV, MERS-CoV, and SARS-CoV-2) that pose significant threats to human health are currently believed to have come from bats. The intermediate host of SARS-CoV is the civet, while MERS-CoV is a camel. The intermediate host of SARS-CoV-2 has not been determined, so there is a concern that the disease is caused by direct consumption of wild animals. So why can't the virus be transmitted directly from bats to people? This is mainly because bats mostly live in areas far from human activities so that viruses need to be passed on by other animals that have close contact with humans. Secondarily, viruses also need to be recombined in other species to be more effective at infecting humans.
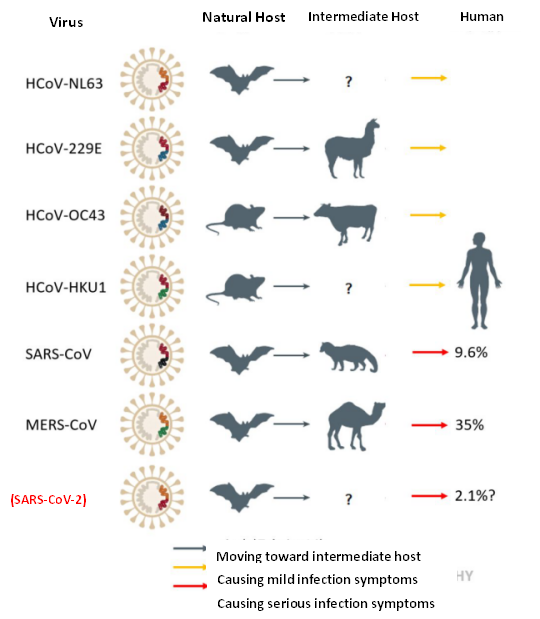
Figure 2. Host migration of pathogenic Coronavirinae (Fung & Liu, 2019)
The Structure of Coronavirus
In order to deal with the enemy, we need to first understand its equipment. We need to know whether there are some weaknesses the virus has in its inherent structure. The most obvious feature of coronaviruses is the protrusions on the edge (resembling a crown), these protrusions are called spike (S) glycoproteins. They function like a key, which bind to the lock (membrane receptor) on the cell membrane and opens the door to the inside of the cell. The spike glycoprotein structures are synthesized by the S gene on the viral genome, and are connected to the lipid membranes. The viral membrane not only facilitates entry into host cells via fusion with cell membranes, but also plays a role in protecting viruses from the immune system.
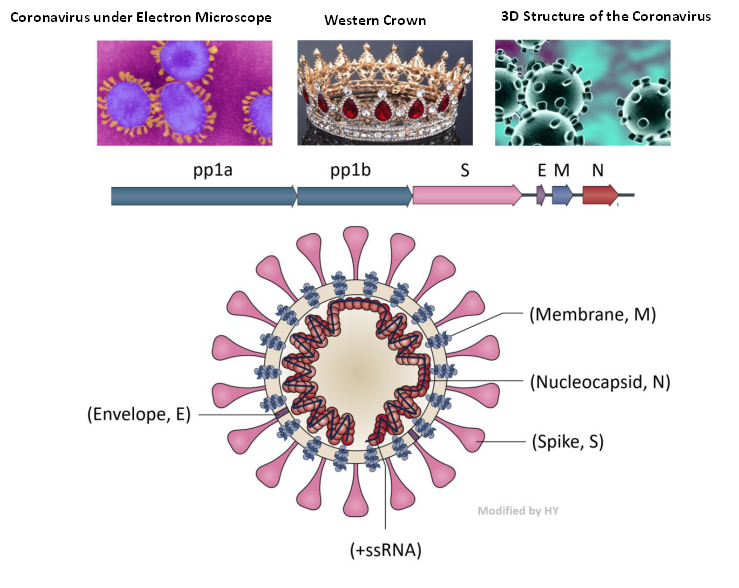
Figure 3. Structure of the Coronavirus
(Jie Cui et al., 2019 and https://www.vice.com)
The next layer is the envelope of the virus. Different from the membrane, this layer is composed of protein and also plays a role in protecting the virus. Alcohol (75%) is able to kill the new coronavirus because it denatures the protein in this envelope, thereby destroying the virus.
Inside the envelope, there is RNA and the nuclear protein that binds with RNA to maintain stability of the virus - the core of the virus. Except for the existing membrane structure, other viral structures need to be synthesized inside the cell. The +ssRNA chain of the coronavirus is unique in that it can directly guide the synthesis of proteins by ‘hijacking’ the cell machinery. In the figure 3, pp1a and pp1b encode about 16 enzymes relate to replication process. The length of these two open reading frames accounts for about 2 / 3 of the total RNA of the virus, and other structural proteins account for about 1 / 3.
The SARS virus of 2019 has the closest relationship with SARS-CoV-2 among all virus that can infect humans, with 79% genomic sequence similarity. In addition to the whole genome comparison, scientists also compared the RNA sequence within the two important open reading frames separately, and found that the spike protein of the 2019 coronavirus is quite different from that of the 2002 SARS virus. However, the latest research shows that the new coronavirus probably entered the bronchial epithelial cells and alveolar cells through its spike protein binding to the cell surface receptor ACE2 – the same target protein that mediates cellular entry for SARS. According to studies of SARS virus, the spike of the coronavirus is mainly composed of S1 and S2 subunits, in which S1 is responsible for the combination with ACE2, and is also the target for vaccine development, while S2 subunit is responsible for the internalization of the virus into cells.
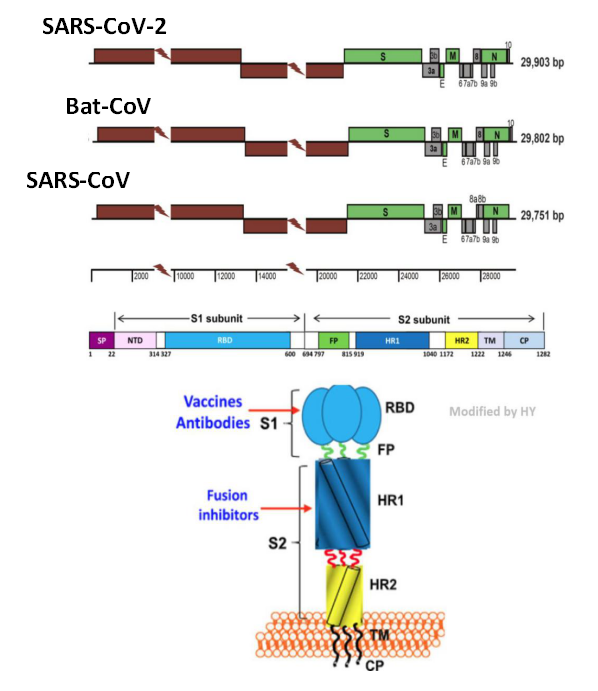
Figure 4. Genome of Coronavirus], and Spike Gene & Spike Protein Structure
(Wu et, al. 2020, Chan et, al. 2020 and Jiang et, al. 2020)
The reason why the virus is difficult to eliminate is that they evolve very fast, especially those that use +ssRNA as genetic material. Such +ssRNA viruses lack the stability of DNA double helix and also undergo recombination with host RNA - making their mutation frequency very high. Although most of the mutations are harmful for the virus, this is solved by the species’ extremely fast replication speed: as long as there is a single mutation that can adapt to the development direction of the environment, the mutated virus will rapidly proliferate.
How do coronaviruses invade cells?
Coronavirus infection in human cells are divided into seven steps:
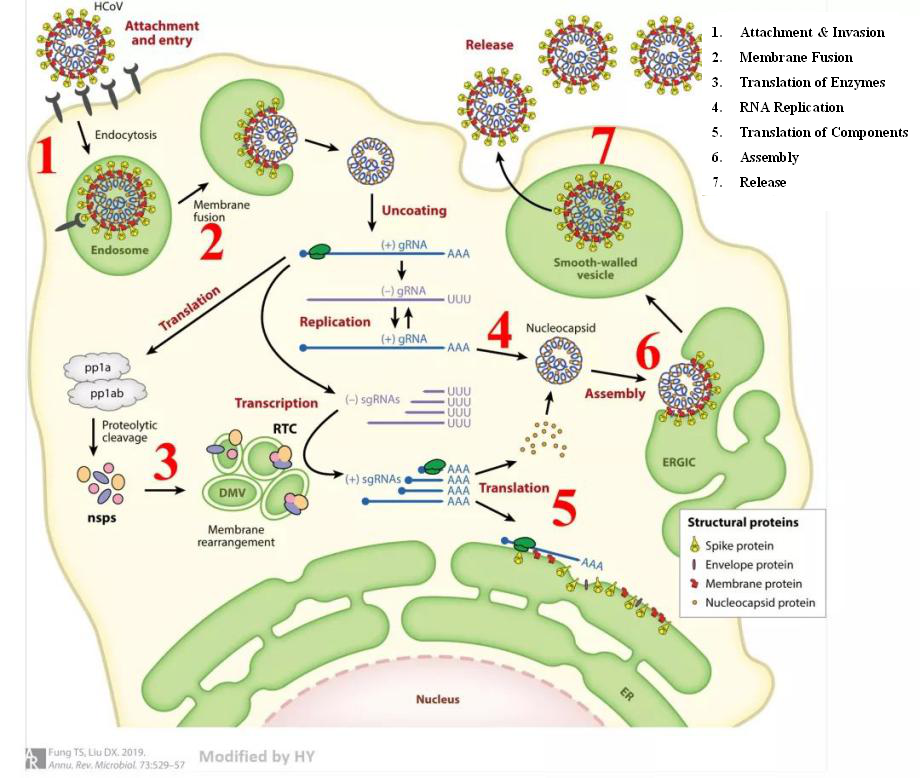
Figure 5. Path of coronavirus invasion and replication (Fung & Liu, 2019)
What treatments can be adopted in curing the novel coronavirus?
The research on the infection mechanisms of SARS-CoV-2 is just beginning, but due to its structural similarities with other coronaviruses, its invasion process could be similar to other pathogenic human coronaviruses. We can design drugs that target each of the seven steps of viral invasion. Regardless of which step blocks the progress of the invasion, the disease will be controlled. The current antiviral methods are mainly divided into three categories: serum/antibody therapy, vaccine therapy, and chemical drug therapy.
Serum / Antibody Therapy
Theoretically, antibodies acting against the new coronavirus can be produced by many patients who are recovering from the invasion of SARS-CoV-2. Will it work to just extract antibodies from recovered patients’ blood? Unfortunately, the answer is a clear “No.”
(1) The components of serum are complex: antibodies produced by the human body are matched with its own immune system. If the antibodies are to be used on others, it will cause a dangerous rejection reaction;
(2) Since the human body is not a bioreactor, there is not much of this kind of antibody in the human body. What’s more, after successfully clearing the virus, the antibody will be reduced. Additionally, there will be some loss of the antibodies in the extraction process, so the antibodies from recovered patients are not enough;
(3) Monoclonal antibodies (mAbs) have a long production cycle that contribute to their high cost - so it is not realistic to use it widely for curing existing patients.
Vaccine Therapy
Smallpox has been eradicated by vaccine efforts on a worldwide scale. While vaccines are good at dealing with relatively ‘stable’ DNA viruses, some fickle ssRNA viruses are not so easily dealt with by the same approach. Smallpox virus has a highly-stable double-stranded DNA structure, which is difficult to change rapidly, making them more easily eliminated by human beings. Since the principle of vaccine is to inject antigen to cause an immune response in vivo, a vaccine can only simulate a virus with the same antigenic determinant. If the viral structure is constantly changing due to mutation, both the antibodies and memory cells synthesized through the previous vaccination will be useless. Moreover, the development cycle of new vaccines is too long to cope with the current time-sensitive situation.
Chemical Drug Therapy
In consideration of both time and current research progress, the best treatment option would be to pursue drug therapy. For drug development, it is better that drugs are able to recognize the specific structure of the virus without affecting the normal proteins in healthy host cells. In order to not affect the overall host, we can also use drugs to weaken the proteins associated with the virus, so as to inhibit its proliferation.
Till now, the known coronavirus has ‘hijacked’host cells to provide internal services for the virus in the seven steps of invasion. The table below summarizes the ‘traitor’ host proteins from all known human-infecting coronaviruses (hCoV) preceding the outbreak of the novel coronavirus (SARS-CoV-2); the red boxes indicate host proteins associated to strongly-pathogenic strains.
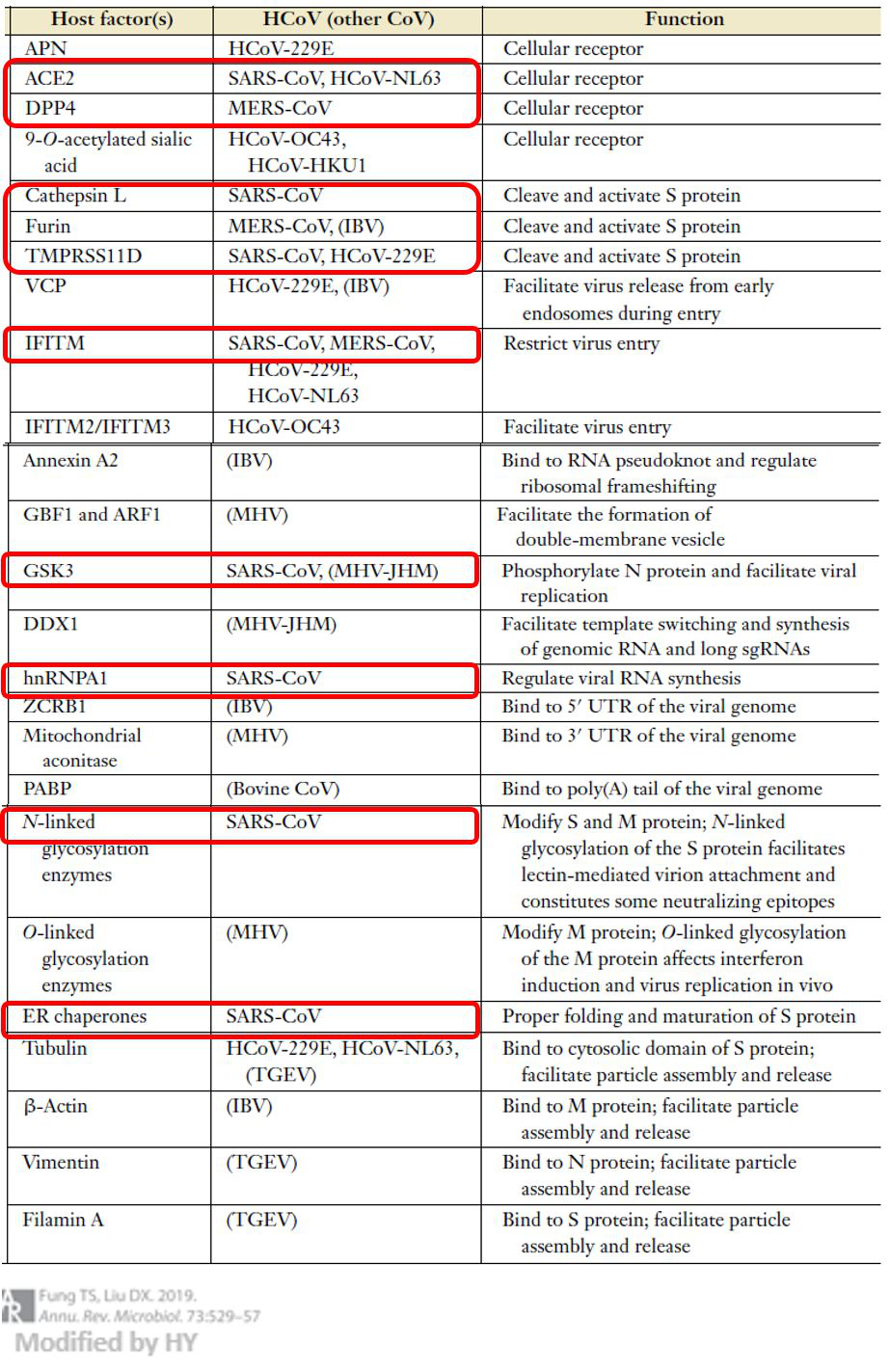
Figure 6. Coronavirus invasion related host protein (Fung & Liu, 2019)
The host factor(s) noted could be potential targets for SARS-CoV-2 antiviral drug research, although we can only infer a limited amount from the closely-related SARS-CoV. Before we clearly understand the invasion mechanism of the SARS-CoV-2, we can use the known pathways of SARS-CoV to quickly identify targets. More importantly, these proteins are required for normal human life activities, so the basic research on the infection mechanism of SARS-CoV-2 is imminent.
Among these potential targets, host cell surface receptors are of particular importance. This is because once the virus molecules enter the cell, the required drugs also need to be able to enter the cell - undoubtedly increasing the difficulty of drug design, given that many biomacromolecule drugs (such as antibodies) are unable to enter the cell. The activity of the virus inside the cell mainly depends on the proteins and energy provided by the host cell, so the side effects of many inhibitors are also of significance. Therefore, drug research and development aimed at membrane surface receptors is the best way to help cells directly 'keep the virus out of the gate'.
Current studies have found that ACE2 is related to the infection of both SARS-CoV-2 and SARS-CoV, and DPP4 is related to the infection of MERS-CoV. At present, APN is only found to be related to the infection of HCoV-229e coronavirus - but this virus will not cause serious disease, only mild cold symptoms. Although hCoV strains combined with APN do not yet cause lethal disease, the expression level of APN in the pulmonary tissue is higher than that of ACE2 (just as MERS virus receptor DPP4). Once the human body infected by the new virus, it may cause lethal respiratory system disease as serious as MERS if it utilizes the APN host factor. What's more, the spike protein coding gene of coronavirus with single stranded RNA is very easy to mutate, which can change its target at any time. Therefore, continued research and drug development for these three kinds of receptors is of great significance - not only for the control of the current epidemic situation, but to also obtain the knowledge that will arm us against potential future outbreaks.
How can we respond to the current epidemic and prevent further outbreaks?
Most viruses in nature are quite specific, generally infecting only a few species; the most successful viruses are not life-threatening for hosts. However, if different species have indirect and frequent contact with a virus, it may lead to the exchange of genetic material between the virus and the potential hosts. These exchanges result in viral adaptation to new host species or lead to more severe infection, bringing unpredictable effects.
There are several lessons learned from this disaster, but everyone can do their part to reduce the possibility of unknown viral infections by avoiding wild animals without adequate safety precautions. We should also increase the investment in medical research, especially in basic research to provide powerful antiviral tools and sufficient logistical technical support for medical workers fighting at the front lines. In addition to retroactively responding to the epidemic, we should also focus on the prevention of future viral outbreaks.
Now with the rapid development of deep learning and artificial intelligence (AI), we can use these techniques together with the large number of virus genome data and mutation data accumulated in the existing research. If we can analyze the virus’ sequence, predict the probability of host migration that may be generated by viral mutation, and accurately construct the 3D conformation of its spike protein as well as other key structures in advance – the potential acceleration of drug design will improve our prospects in dealing with the virus. Anticipating the viral enemy first will minimize the human harm caused by the virus, and even kill the virus before it infects our species.
Postscript
Whether it is vaccine development, new drug development, or gene therapy, we need accurate animal models to verify the pathogenesis and immune mechanisms of diseases such as COVID-19. As a mature animal model service provider, Cyagen’s R&D experts have been doing their best to develop animal models related to SARS-CoV-2 research to contribute to the global outbreak prevention effort.
Coronaviruses invade human cells mainly through binding to membrane receptors; there are three cellular receptors are related to hCoV infection: ACE2, DPP4, and APN.
Although the APN receptor is not associated with lethal disease, its high expression levels in the lungs (similar to DPP4) make it an ideal viral target to cause respiratory system diseases as severe as MERS once human body is infected. The S gene of coronavirus, which encodes the spike proteins that bind with cell surface receptors, undergoes high mutation rates that could change virus affinity towards target receptors – allowing it to enter the cell in a new manner altogether, which could be seriously life-threatening. For these reasons, the biological expert team from Cyagen carried out the development of gene modification models for the three kinds of key receptors of coronavirus at the first opportunity – our way of contributing to the research on prevention and control of the epidemic.
References:
We will respond to you in 1-2 business days.