
A recent study has suggested that that NF-kB activation is boosted by colocalization of engaged immune receptors (i.e., CD40) with RAB7 small GTPase on mature endosomes in mouse B cells. This research was enabled by using a conditional (floxed) gene knockin mouse model - Cd19+/creRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) – which is designed to express untagged RAB7 in CD19+ B cells. Given that the Rab7 B-Tg mouse model is a promising tool for continued studies of RAB7A and NF-kB activation, the prerequisite TurboKnockout®-generated Rosa26+/fl-STOP-fl-Rab7 (a.k.a. Rosa26fl-Stop-fl-Rab7) strain will soon be available to researchers through JAX Mice.
Herein, we cover a general background of RAB7A, the importance of the Rab7 B-Tg mouse model, and an outline of the research performed to-date.
The human RAB7A (previously known as RAB7) gene encodes Ras-related protein Rab-7a (RAB7A), which is a member of both the Rab protein family and the Ras superfamily of small G-proteins. Small G-proteins, also knock as small GTPases, are a family of hydrolase enzymes that can hydrolyze guanosine triphosphate (GTP) upon binding.
RAB family proteins are peripheral membrane proteins which serve to regulate many steps of both membrane traffic and vesicular transport. Each RAB protein targets multiple proteins involved in a variety of endocytic and exocytic pathways – influencing the routes through which proteins are moved in the cell. The RAB7a protein regulates vesicle traffic in the late endosomes and traffic from these to the lysosomes. Mutations of highly conserved amino acid residues in RAB7A have caused some forms of Charcot-Marie-Tooth (CMT) type 2 neuropathies. An important paralog of RAB7A is RAB9B.1
Associated Diseases: Charcot-Marie-Tooth Disease, Axonal, Type 2B and Charcot-Marie-Tooth Disease
Related pathways: RAB GEFs exchange GTP for GDP on RABs and TBC/RABGAPs. The featured case study has shown that RAB7 is an integral component of the B cell NF-kB activation machinery
Common gene aliases:
The Rab7 B-Tg (Cd19+/creRosa26+/fl-STOP-fl-Rab7) mouse model was developed to address how endosomal RAB7 mediates NF-kB activation for B cell differentiation in vivo – numerous factors were incorporated in the model design process, with the key details as follows. Considering that surface receptors (e.g., CD40 or TLR4) may be internalized upon engagement in several immune cells, a subset of endocytosed receptors would reach RAB7-marked endosomes and transduce signals from there. Additionally, investigators postulated that the lipid raft-dependent pathway would supplement RAB7-dependent pathway in NF-kB activation when the expression/activity of RAB7 is inhibited. Thus, researchers aimed to explore the mechanistic basis of RAB7-dependent pathway in NF-kB activation by increasing the contribution of this pathway through two complementary strategies:
In the study, these approaches are supported by comprehensive approaches in molecular biology, cell biology, imaging, and in silico docking to analyze receptor internalization, their colocalization with RAB7 and TRAF6, NF-kB activation in single cells, and induction of activation-induced deaminase (AID) and class switch DNA recombination (CSR). Together, these two strategies have revealed an important role of RAB7- dependent “intracellular membrane signalosomes” in promoting TRAF6 K63 polyubiquitination and NF-kB activation during B cell differentiation.
The Rab7 B-Tg conditional (floxed) gene knockin mouse model is designed to express untagged RAB7 from the Rosa26 locus (which is also close to the endogenous Rab7 locus, Fig. 1). This genetic engineering & breeding approach resulted in increased Rab7 transcript levels and RAB7 overexpression in CD19+ B cells. Notably, the development, maturation, and survival of B cells were all unaffected in Rab7 B-Tg mice.
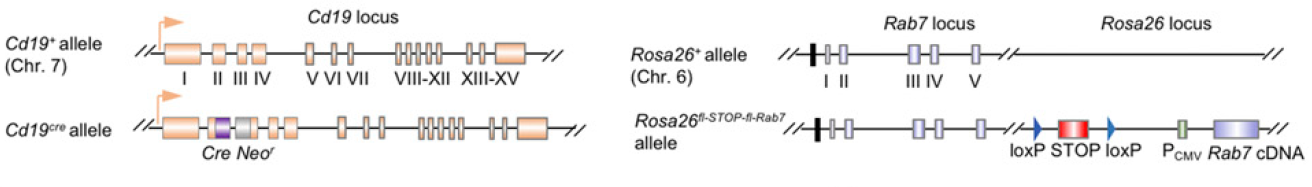
Figure 1: Illustration of Cd19 and Rosa26 alleles in Cd19+/creRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) mice.
The Rab7 B-Tg mouse model is achieved by the following methods, as described in the publication (emphasis ours):
“Cd19+/creRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) mice and their wildtype littermate controls were generated by crossing Cd19+/cre mice (The Jackson Laboratory, stock number 006785) with Rosa26+/fl-STOP-fl-Rab7 mice, as generated by a customized TurboKnockout approach (Cyagen Biosciences). Briefly, the targeting vector contains two arms homologous to the first and second exon, respectively, of the Rosa26 locus. In between the arms lies the CMV promoter-driven Rab7 cDNA, which is preceded by a loxP site-flank “STOP” cassette sequence, as followed by a neomycin resistance gene (Neor) in the inverse direction (Fig. 1A). After injection of the targeting vector into embryonic stem cells from C57 mice and screening by Southern blotting, selected embryonic stem cell clones showing correct targeting were used for blastocyst microinjection to produce chimeric mice. These were bred with a C57 mouse strain that expressed Flippase to screen for mice with germline transmission as founders; as Flippase also deleted Neor in the offspring, the entire process to generate founders on the C57 background with no Neor took only 6 mo. Among the five founders (one female and four male heterozygous mice), two were randomly selected for breeding with Cd19+/cre mice to establish independent lines. As progenies from these two lines displayed no significant difference in the parameters we measured, data from one line are depicted in this study. In Rab7 B-Tg mice, Cre expression driven by the Cd19 promoter in the Cd19Cre allele mediated deletion of the STOP cassette in the Rosa26 locus, thereby allowing for the CMV promoter-driven expression of untagged RAB7 in CD19+ B cells.”2
The Rosa26+/fl-STOP-fl-Rab7 (a.k.a. Rosa26fl-Stop-fl-Rab7) founder mice were generated in just 6 months due to Cyagen’s innovative TurboKnockout® Gene Targeting approach. With TurboKnockout®, the self-removing Neor selection cassette circumvents the need to breed to Flp deleter mice, accelerating the model development process. These same innovations may be used to generate custom strains to study other genes of interest, saving time and avoiding the difficulties of breeding procedures. In addition to developing simple point mutation and knockout (KO) mice, the TurboKnockout® approach has been used to develop conditional ready (e.g. floxed) and humanized mouse models with large fragment knockin (LFKI) up to 300 kb.
Compared with Targeted Gene Editing-based techniques, Cyagen’s proprietary TurboKnockout® technology is free of patent disputes, provides precise models on a comparable timeline, and is the method of choice among our researchers performing drug development projects.
Given the potential applications for the Cd19+/creRosa26+/fl-STOP-fl-Rab7 (Rab7 B-Tg) mice, Dr. Zhenming Xu has chosen to donate the requisite Rosa26+/fl-STOP-fl-Rab7 (a.k.a. Rosa26fl-Stop-fl-Rab7) mouse strain to the JAX Mice repository.
Produced with Cyagen’s customized TurboKnockout® approach, Rosa26fl-Stop-fl-Rab7 mice express full-length mouse Rab7 cDNA upon cre recombinase-mediated excision of a loxP-flanked Stop cassette. Crossing this strain with Cd19+/cre mice generates Rab7 B-Tg mice, which had B cell-specific overexpression of RAB7, increased antibody responses, coexistent with increased activation-induced cytidine deaminase (AID) expression and class switch DNA recombination (CSR). At the time writing, the Rosa26fl-Stop-fl-Rab7 mice are estimated to begin distribution from JAX Mice on March 29, 2021.3
The referenced study developed the Rab7 B-Tg mice to demonstrate that NF-kB activation is boosted by colocalization of engaged immune receptors (e.g. CD40), with RAB7 small GTPase on mature endosomes in mouse B cells. RAB7 directly interacts with TRAF6 E3 ubiquitin ligase in mature endosomes, which catalyzes K63 polyubiquitination for NF-kB activation. Thus, the RAB7 overexpression seen in Rab7 B-Tg mouse B cells upregulates K63 polyubiquitination activity of TRAF6, enhances NF-kB activation and activation-induced cytidine deaminase induction, and boosts IgG Ab and autoantibody levels. Collectively, results show “that RAB7 is an integral component of the B cell NF-kB activation machinery, likely through interaction with TRAF6.”
Cited Publication Details
| Model Generation Approach - Type | TurboKnockout® Gene Targeting |
|---|---|
| Mouse Strain Developed | Rosa26+/fl-STOP-fl-Rab7 (a.k.a. Rosa26fl-Stop-fl-Rab7) |
| Research Applications | Immunology, Inflammation |
| Gene Target | RAB7 |
| Gene Modification Type | Rosa26 Conditional Ready (Floxed) |
| Title | B Cell Endosomal RAB7 Promotes TRAF6 K63 Polyubiquitination and NF-κB Activation for Antibody Class-Switching |
| Journal | The Journal of Immunology PMID: 32277056 |
A multitude of experimental methods were used in the referenced research study - including flow cytometry, intracellular staining, flow imaging, immunofluorescence microscopy, intracellular membrane fractionation, immunoblotting and coimmunoprecipitation, biofluorescence complementation assay, and real-time quantitative RT-PCR transcript analysis. The data collected from these were interpreted by the investigators to signify the following results:
Future studies may verify the role of TRAF6–RAB7 interaction in NF-kB activation by implementing models with point mutations at the interaction interface to determine those which disrupt such activity and lead to CSR impairment. Due to the prolonged length of B cell stimulation (e.g., 24–48 hours) required to reveal of the multi-staged function of RAB7-dependent intracellular membrane signalosomes, the true kinetics of formation may have been missed in previous studies.
Although NF-kB hyperactivation is responsible for a range of disease conditions, NF-kB is a poor target since it is critical in homeostasis. However, RAB7 could be a common causative factor among such NF-kB hyperactivation-related conditions, making it an attractive therapeutic target. The disruption of RAB7-dependent intracellular membrane signalosomes could allow plasma membrane signalosomes to continue providing basal NF-kB activity to maintain essential functions. This may be achieved by interfering with the RAB7-TRAF6 interaction, which may be tested with models that implement point mutations at the interaction interface.
We will respond to you in 1-2 business days.