
With the advantages of simple operation and high recombination rate, the Cre-Lox system has now become a powerful tool for genetic manipulation across in vivo and in vitro. The Cre-Lox system enables gene expression with exceptional spatial (expresses a gene in specific cells, tissues or entire organisms) and temporal (at specific time points through the application of the Cre-Lox system) control. Application of the Cre-Lox system is often used to achieve custom spatiotemporal operations on specific genes, which is useful for the study of gene function and animal models of human disease. Herein, we explore the Cre-Lox system and its profound impact on animal model construction capabilities.
There are two main parts of the Cre-Lox system, both of which have been adapted from the P1 bacteriophage:
Cyclization recombinase (Cre) is one of the tyrosine site-specific recombinases, which is known to catalyse the site specific recombination event between two DNA recognition sites (LoxP sites). Cre recombinase consists of 343 amino acids, which can specifically recognize Lox sites. In addition to Cre, there are flipase (Flp) and D6 specific recombinase (Dre) – although these enzymes still demonstrate a lower recombination efficiency compared with Cre.
The palindromic DNA region recognized by Cre recombinase are known loxP (locus of X-over P1) sites. LoxP sites are directional 34 bp sequences made up of two 13 bp reverse complement on both sides (the recognition sequence of Cre recombinase) and an 8 bp spacer region (the position where recombination occurs) which gives it directionality. The orientation of LoxP sites can result in three genetic recombination events, including excision, inversion, and translocation. In addition to wild type LoxP sites, there are common sites such as Lox2272, Lox511, Lox5171, and more - these mutant Lox sites can also be recognized by Cre recombinase. However, only two of the same mutant lox sites can recombine with each other.
Table 1. Published mutant loxP sites
| loxP site | Left inverted repeat sequence | Spacer (5'→3') | Right inverted repeat sequence |
|---|---|---|---|
| Wild-type | ATAACTTCGTATA | ATGTATGC | TATACGAAGTTAT |
| lox 511 [6] | ATAACTTCGTATA | ATGTATaC | TATACGAAGTTAT |
| lox 5171 [7] | ATAACTTCGTATA | ATGTgTaC | TATACGAAGTTAT |
| lox 2272 [7] | ATAACTTCGTATA | AaGTATcC | TATACGAAGTTAT |
| m2 [8] | ATAACTTCGTATA | AgaaAcca | TATACGAAGTTAT |
| m3 [8] | ATAACTTCGTATA | taaTAcca | TATACGAAGTTAT |
| m7 [8] | ATAACTTCGTATA | AgaTAgaa | TATACGAAGTTAT |
| m11 [8] | ATAACTTCGTATA | cgaTAcca | TATACGAAGTTAT |
| lox 71 [12, 13] | taccgTTCGTATA | ATGTATGC | TATACGAAGTTAT |
| lox 66 [12, 13] | ATAACTTCGTATA | ATGTATGC | TATACGAAcggta |
Compared with constitutively expressed gene mutations, conditional models provide greater temporal and regional control of gene expression. The Cre-Lox recombination system enables the generation of tissue-specific or inducible knockouts with a high level of control over the spatial and temporal expression of genes.
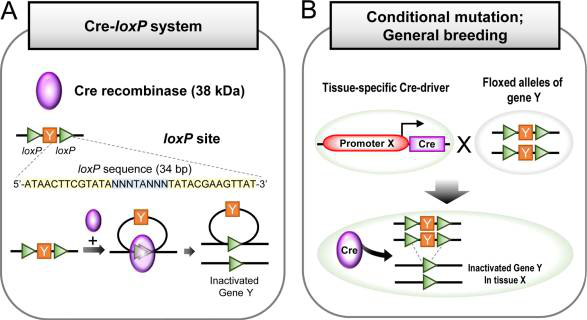
(A) An overview of Cre-LoxP system. 38 kDa Cre recombinase recognizes the loxP sites of specific 34 bp DNA sequences, excising the sequence and inactivating Gene Y. (B) General breeding strategy for conditional mutation using loxP and Cre driving mouse line. Tissue-specific Cre-driver crossing with Floxed alleles of gene Y leads to inactivation of Gene Y in tissues associated with Promoter X.
As outlined in Figure 1 above, the basic Cre-loxP recombination event is most often used for controlled excision of genetic sequences. This has been adapted to create Cre-dependent sequence knockout (KO) – depicted in figure 1 - and Cre-dependent gene expression models – such as the transgene switch strategies described below.
Targeted Gene Editing Capabilities for Mouse and Rat Models:
Cyagen Cre Model Platform
Despite the utility of the Cre-Lox modeling strategy, the complexity of breeding protocol alongside the increased cost of rat colony maintenance has created significant barriers for researchers seeking to make novel conditional rat models. Another major constraint faced by researchers developing cKO or cKI rat models is the lack of Cre lines specific to the target of interest.
As a leader in custom rodent models, Cyagen is building a repository of Cre loxP model resources, including Cre driver rat strains. We are offering a complete range of custom conditional knockout (cKO) or conditional knockin (cKI) rat models alongside the Cre driver strains needed to develop physiologically relevant models of human diseases, available to researchers worldwide.
Below are some of the current Cre rat strains under-development:
| alb(Cre Ert2) | myh6(Cre Ert2) | slc6a3(Cre Ert2) | camk2a(Cre Ert2) |
| alb(Cre) | myh6(Cre) | slc6a3(Cre) | camk2a(Cre) |
| gfap(Cre) | nes(Cre) | nphs2(Cre Ert2) | |
| gfap(Cre Ert2) | nes(Cre Ert2) | nphs2(Cre) |
Still looking for your Cre line? Cyagen can provide pricing accommodations to researchers interested in Cre lines not currently available in our listings. Contact us to inquire about your conditional model needs and see how we can help develop a custom Cre strain to support your field of study.
About Cyagen
Cyagen offers a "one-stop shop" tailored to your gene research and model generation needs. Our services range from DNA vector construction to embryonic stem cell manipulation, microinjection, and breeding. All projects are fully customizable and flexible.
|
|
|
|
|
Photos of the Cyagen Transgenic Animal Center (CTAC) |
||
References:
We will respond to you in 1-2 business days.