
It is well known that genes play a major role in many human diseases - for this reason they are an important research topic in the field of life science and medicine.
How can the public quickly grasp the recent research on disease-related genes, when the only resources out about them are long complex academic journals? To save the time and energy of researchers, Cyagen has launched its new project -‘Gene of the Week’, in which we will introduce one specific gene related to human disease each week.
First gene feature: APP (Amyloid Precursor Protein)
Background Information
1. Human APP Gene
Gene location:chromosome 21 (21q21.3)
Full length: spans 290 kb and includes 18 exons
Values of amino acid: 639-770AA
Conservation: Nematodes, drosophila, and all vertebrates
Cleavage product: sAPPα, sAPPβ, Aβ, C83, C99, AICD, P3
Cell location: membrane protein
Protein molecular weight: ~87kd
Number of major protein amino acids: 770,695,751
Gene family: APLP1, APLP2
2. Mouse App Gene
Gene location:chromosome 16 (16 C3.3;)
Full length: spans 222 kb and includes 19 exons
Knockout (KO): prolong the long-term enhancement; affects learning and memory; both the App gene knockout (KO) and conditional knockout (cKO) mouse models are available in our Mouse Model eBank.
Overexpression: Long-term enhancement and weakening
Common models: 5×FAD, 3×Tg, APP/PS1, APPswe
3. Rat App Gene
Gene location:chromosome 11 (11q11)
Full length: spans 217 kb and includes 18 exons
Knockout (KO): no validation data
Overexpression: no information
Common models: APP21, APPKI, APPPS1
Overview of APP Gene Research
The main physiological functions of the amyloid precursor protein (APP) have not been fully studied, but some of its mutations can lead to an increase in Aβ (Amyloid beta peptide) production, or an increased likelihood of Aβ aggregation. The accumulation of Aβ (whether it is oligomers or senile plaques) can lead to disturbances in cellular calcium signals and mitochondrial function, which in turn contributes to the loss of synapses and neuronal death, as well as a neuroinflammatory cascade - which is currently the leading Aβ hypothesis. Another study believes that Aβ is a byproduct of Alzheimer’s Disease (AD), which has a protective effect on neurons in the early stage of AD. These hypotheses are discussed in a previous article:“Applications of Rat Models in Alzheimer's Disease Discovery Research.”
Normally, APP is cut by an α-secretase enzyme produced by ADAM10, which cuts Aβ in a way which prevents the formation of neuritic plaques from accumulation of Aβ. This is known as the non-amyloidogenic pathway (α-pathway), that can be described as a "good" cleavage that does not release toxic Aβ. However, if APP is first cut by β-secretase (BACE1), the amyloidogenic cascade pathway (β-pathway) is initiated. The overactive β-pathway will cause Aβ overload and lead to formation of amyloid precipitation and neuritic plaques – indicating onset of AD.
In many familial AD cases, APP mutations can be roughly divided into three categories:
In addition, the activities of enzymes that are involved in Aβ clearance such as IDE, NEP, ECE, ACE, MMP, etc. , have also been found to be reduced in AD-afflicted tissues. In recent years, some mutations in APP have also been found to reduce the incidence of AD.
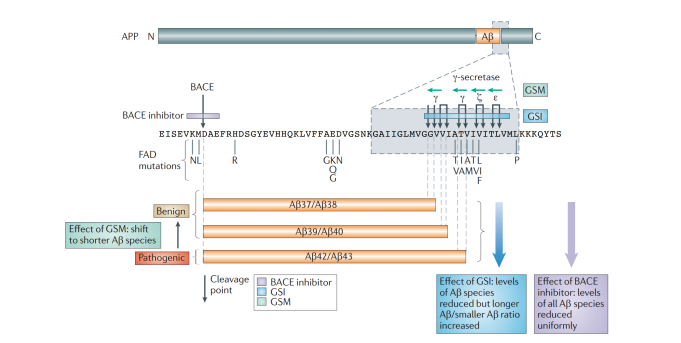
Figure 1. APP mutation and cleavage
Alzheimer's disease (AD) is one of the most common neurodegenerative diseases in the world. The extracellular amyloid plaques and intracellular neurofibrillary tangles characterize the clinical manifestations of Alzheimer's disease, which results in neuronal dysfunction and cell death.
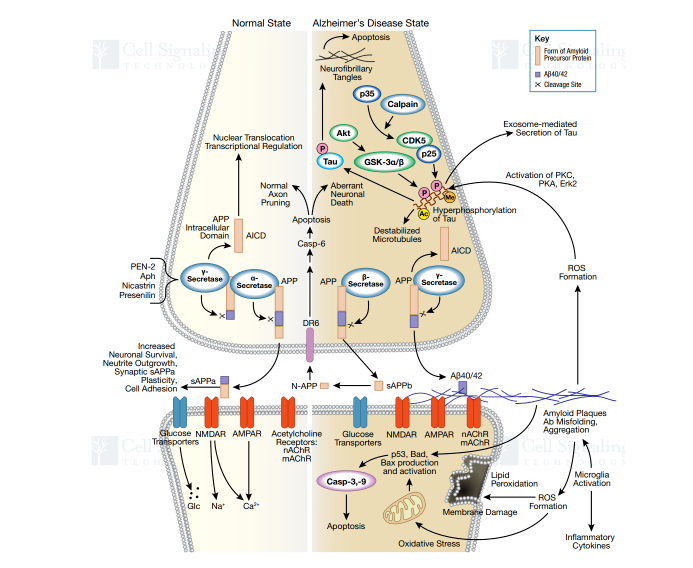
Figure 2. Signaling Pathway of AD
APP Expression in Human Tissues
APP is highly expressed in the brain tissue, thyroid, and kidney of adults. However, the expression abundance is low in other major organs and tissues of digestive tract.
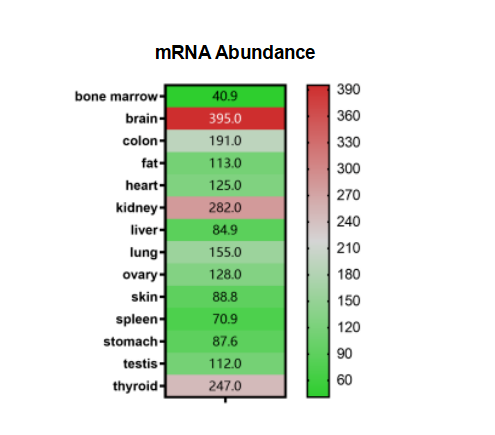
Figure 3. APP Expression (relative expression values)
References:
DOI: 10.1146/annurev-neuro-070815-014015. Epub 2016 Apr 4. PMID: 27050320.
We will respond to you in 1-2 business days.