
Alzheimer's disease (AD) is the most common age-related neurodegenerative disorder, which is affecting 20 to 30 million individuals worldwide now. Currently, no effective therapies to treat or cure this disease were developed. In this review, we discuss the current research and understanding of the PSEN1 gene and its mutation in causing familial Alzheimer's disease (FAD).
Background Information - PSEN1 & Homologs Across Species
1. Human PSEN1 Gene
Gene location:Chromosome 14 (14q24.2)
Full length: Spans 87kb and includes 14 exons
Values of amino acid: 467AA
Conservation: Nematodes, fruit flies, and all vertebrates
Cleavage product: Notch receptor, APP
Cell location: membrane protein
Protein molecular weight: ~53kd
Gene family: PSEN2
Related diseases: Alzheimer's disease (AD), Pick's disease (FTD, frontotemporal dementia)
2. Mouse Psen1 Gene
Gene location:Chromosome 12 (12D1; 12 38.84cM)
Full length: spans 47kb and includes 12 exons
Knockout (KO): Nerve regeneration is impaired, and loss of neurons
Overexpression: No significant phenotype
Common models: 5×FAD, 3×Tg, APP/PS1
3. Rat Psen1 Gene
Gene location:Chromosome 11 (11q11)
Full length: Spans 217kb and includes 18 exons
Knockout (KO): No validation data
Overexpression: No information
Common models: APP21, APPKI, APPPS1
Overview of PSEN1 Gene Research
PSEN1 is gene associated with familial Alzheimer's disease that encodes the Presenilin 1 protein. Some mutations of this gene can cause the increased production of the toxic, longer form of amyloid-beta (Aβ) fragments to be initiated. The protein encoded by this gene is the active part of gamma- (γ-) secretase, a multi-subunit protease complex is involved in the cleavage of Notch receptors, with PSEN1 participating in the release of synaptic transmitters in hippocampal vertebral neuritis. This suggests that presenilin 1 plays a critical role in the Notch signaling pathway.
Currently, there are 318 mutations of PSEN1 that have been identified, but more are added every year. Of these mutations, around 300 are linked to Alzheimer's disease, and some are related to other neurological diseases such as Pick's disease, frontotemporal dementia, Amyotrophic lateral sclerosis (ALS), and more.
Figure 1. The Distribution of PSEN1 Mutation
(Source: https://www.alzforum.org/mutations/psen-1)
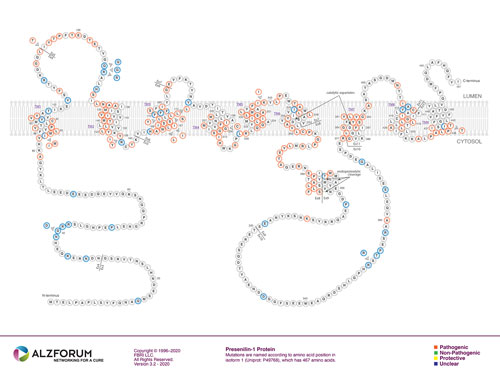
(Left) N-terminal sequence; (Right) C-terminal sequence; Red circle - pathogenic mutation; Yellow - protective mutation; Blue - unknown phenotype of the mutation; Arrows indicate possible deletion or insertion mutations. The proteins encoded by different exons are separated by lines.
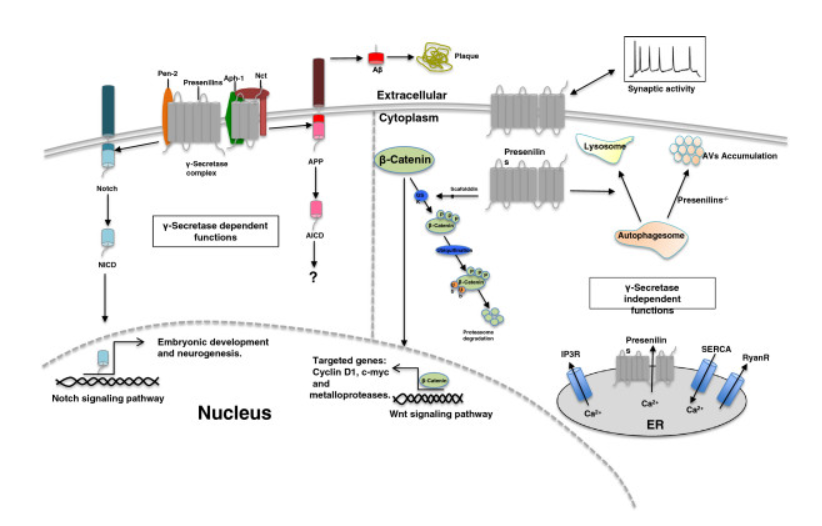
The function of PS1 (Presenilin 1) can be divided into γ-secretase-dependent and γ-secretase-independent regions. Current research shows that both membrane receptors amyloid precursor protein (APP) and Notch are cleaved by the γ-secretase. PSEN1 alone can stabilize β-Catenin in the Wnt signaling pathway, serving as a negative regulator in Wnt signaling. Calcium signaling in related pathways is regulated by the combination of original calcium ion (Ca2+) channels or as Ca2+ channels independently functioning in the endoplasmic reticulum (ER). The lysosome-associated functions during autophagy are all regulated by PSEN1, and the phenotype also regulates the plasticity of synapses.
Figure 2. Signaling Pathway of PSEN1[1]
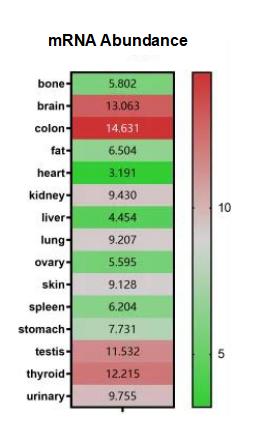
PSEN1 Expression in Human Tissues
PSEN1 is highly expressed in brain, rectum, testes, thymus, and urinary system. However, the expression abundance is low in fat and bone tissues, as well as in the heart, ovaries, spleen, and stomach.
Figure 3. PSEN1 Expression (relative expression values)
Additional Resources
As AD research has progressed, there has been a large movement towards the use of rat models, mainly due to drug development efforts centered on the amyloid hypothesis failing in clinical trials (wherein mouse models were used). Our experts have put together an article describing the advantages that rats may provide in researching AD and related neurodegenerative disorders. View the full text: Applications of Rat Models in Alzheimer's Disease Discovery Research.
References:
We will respond to you in 1-2 business days.